OK, just kidding. But this post involves Caltech dorms, and I feel I have to take part in some old-fashioned school rivalry. There is, in fact, only one college dorm worth talking about.
Anyway, a few years ago, a paper came out that was studying the structure of Facebook social networks on some college campuses:
Traud, A., Kelsic, E., Mucha, P., and Porter, M. Comparing Community Structure to Characteristics in Online Collegiate Social Networks, SIAM Review, 2011, Vol. 53, No. 3: pp. 526-543
In 2011, this Facebook dataset was used in a visualization competition that Conrad Lee described in a post on his blog Sociograph. You can see some of the results in the post; perhaps not surprisingly, the visualizations were hitting the hairball ceiling.
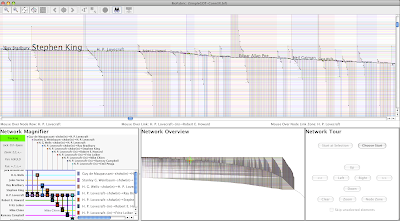
In another posting on Sociograph in late 2012, Conrad used the Caltech portion of that Facebook dataset to illustrate how to visualize adjacency matrices in Python. With that dataset, I've visualized the Caltech Facebook network using BioFabric. With 769 nodes and 33312 16656* edges the network is very high aspect ratio (43:1). I have 15,000 pixel-wide version of the network that you can scroll back and forth with here on my blog, and in my initial iteration of this post, I embedded the file directly on this page. But at about 3.8 MB, it's a little hefty to have to download it when you first visit the blog. So I've included just a detail snapshot below, and you should go to the special scroll page to view it now:
(Note the students' names in the data were anonymized to numbers.)
Since this network needed to be preprocessed a bit to get the final layout, and since it uses a feature I have not yet talked about (link groups), I'll spend this post talking a little bit about I built it.
In addition to providing the links, the data set also indicates the House affiliation (i.e. dorm) of each student (there are eight dorms), and this turns out to be an important aspect of this network. So let's use that data and have BioFabric show the clusters. As I have pointed out before, it's not yet a built-in BioFabric 1.0 feature to automatically do clustered layouts, (though I am working on it!), so some basic scripting is needed. I'm not going to get into the low-level detail of showing the scripts, but just give a high-level description of the steps involved.
First, using the dorm assignments, we identify which edges are in-dorm (Facebook friends in the same dorm), and which edges are between-dorm (Facebook friends in different dorms). Then, using just the in-dorm links, we create eight separate SIF files of in-dorm links, one per dorm. Separately loading each into BioFabric, we can get eight per-dorm BioFabric default layouts (i.e. we are going to use BioFabric to handle the default layout step, instead of scripting it as well). The resulting node orders, which we will use to create a single global ordering file, can be simply exported, just choose Select File->Export->Export Node Order:
 |
| Click on image to enlarge |
(As a side note, the Export Link Order option in that menu is the best route to seeing how to create the edge attribute files you need to explicitly layout edges).
Since we want to order the dorms from biggest to smallest, number the eight dorm node order files in that fashion, e.g. dorm1.noa (biggest) to dorm8.noa (smallest). You'll also need to chop off the first line of each of these files, using e.g.:
tail -n +2 < dorm1.noa > dormr1.noa
Then, to create the single global node ordering file, just do this on the Unix command line:
cat dormr*.noa | awk '{print $1 " = " NR-1}' | sed '1 i Node Row' > globalOrdering.noa
That takes care of specifying the node ordering we will need. At the same time, we want to create the single full-network SIF file where each link is tagged with a suffix indicating whether is it in-dorm (tagged -ic, for in-cluster), or between-dorm (tagged -bc, for between-cluster). We were figuring that out above when we created the eight separate dorm-only networks, so also use that information to tag the links to write out the final single SIF input file.
Then, import the global SIF file, and after the network is loaded, re-layout the whole network by specifying node order. Just select Layout->Layout Using Node Attributes..., use the globalOrdering.noa file you generated, and the network now has the eight separate dorms broken out.
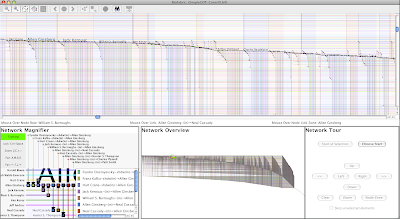
When a network gets long and thin like this, I'm quick to turn on shadow links to get a better idea of what's going on. Just select Edit->Set Display Options... and check Display Shadow Links box. At the same time, I like to shade the node zones, so also check Node Zone Shading before clicking OK. This allows you to see all the Facebook connections for a student by just looking at the node zone for that student.
There is now one more step. As it currently sits, each student has a single edge wedge for all his/her Facebook friends. The tiny subnetwork of three students shown below illustrates that. Although the links going to the node lines right above and below these students correspond to the in-dorm links, that distinction is completely hidden:
 |
| Click on image to enlarge |
So we want to separate the links into the in-dorm (-ic) and between-dorm (-bc) groups so we can see separate edge wedges for these two sets. Since we tagged the links in the SIF input with suffixes, we can easily use that information to create the two distinct edge wedges. Just go to Layout->Specify Link Groups...:
 |
| Click on image to enlarge |
In the dialog, click Add New Entry... twice and enter in the two groups, -ic and -bc:
 |
| Click on image to enlarge |
Click OK, and the network is laid out. Because of the link grouping, we can now easily visualize the two -ic and -bc classes of links for each student. Compare this version below with the one above. The first diagonal for each student are the in-dorm -ic links, as that was the first link group we specified. As expected, those links end at the node rows near these students, i.e. the other nodes in the same dorm. The following single edge wedge of -bc links over on the right side of each node zone tends to look like two separate wedges, since they are connecting to both the dorms above and below this dorm:
 |
| Click on image to enlarge |
Note how the grouping lets us instantly see which students are mostly in-dorm focused with their Facebook connections (e.g. 590), and which have more connections outside the dorm (e.g. 20).
That's it for details on building the network. So go back and have a look at the whole network in the 15,000 pixel-wide version up at the top of this post. You can see the eight separate runs of dorms, compare the two different types of connections, and get an idea of how the students interact.
My next post or two will cover a couple of interesting aspects of this network, but it will be awhile, as I'll be on the road next week to the ISMB/ECCB 2013 conference. If you happen to be there, come say hi at my BioFabric Birds of a Feather session on Monday, July 26th!
* Correction: The original edge number of 33312 that I gave did not account for the equivalent reverse edges in the SIF file of the undirected graph getting thrown out on import to BioFabric. Though the view is actually showing 33312 edges in it since shadow links are turned on.